What Type of Medical Imaging Data Exists?
common file types, open data sets, and the information we store about the human body
last update: Jan 30, 2025
Article Motivation
- What are the common file types in medical imaging?
- What information is saved in those files?
- Where can I find large amounts of good, free data?
- What type of data can we obtain about the human body?
Intro
Every medical imaging modality measures and stores physical parameters of the human body.
2D x-ray stores the total attenuation factor (how much the tissue absorbs the radiation) of all the tissues in our body along the axis of the propagation of the x-rays. The modern way of collecting this data is on a 2D array of photo-diodes (see fig. 1). We could think of the data we need to store as a 2D array of scalar values.
Around the globe it is still very common to take x-rays sheets of film. This analog option can produce great images, but since we are more interested in the digital data that we can obtain, we won't focus on that here.
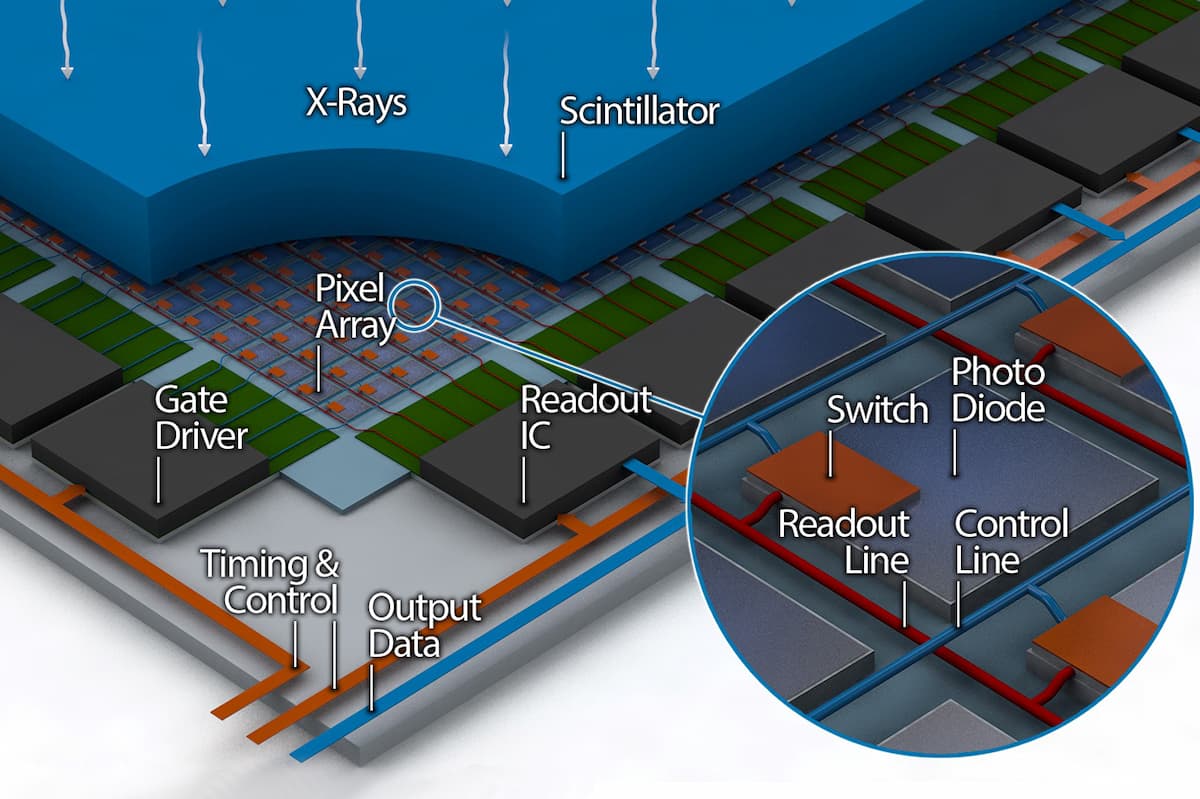
Figure 1: A diagram of a 2D x-ray detector. The data is collected as a 2D array of photo diodes called Pixel Array in this image.
A computed tomography scan (also known as CT or CAT scan), also measures the attenuation values and stores the raw data as an array. MRI typically measures T1 and T2 relaxation time of tissues. Ultrasound measures the intensity and timing of reflected of acoustic signals (electrical signals really, since the source and receiver is a piezoelectric device).
All of these modalities produce raw data that can be reconstructed into image. All medical imaging needs more then just the image file. It needs to include a bunch of meta-data about the image. Things like patient name, height, weight, image type, study date, study time, modality, device manufacturer, scan settings, medical codes, slice thickness, etc. etc. That is where DICOM files come in.
DICOM
DICOM its an international standard for transmitting and storing medical image data. It also is a file type (.dcm).
The intellectual property of DICOM is owned by The National Electrical Manufacturers Association (NEMA) which is a trade association of 350 companies including Canon, DuPont, Duracell, Eli Lilly, GE, Mitsubishi and more.
DICOM grew out of a need for hospitals, doctors offices, medical specialists, and imaging centers to be able to communicate medical images in some uniform way.
The standards of DICOM are now a 4100 page book that details everything about the storage and transmission of medical images. It covers data structures, encoding, how it should be sent over the internet, security protocols, and much more.
What does a .dcm file look like?
Lets download one. We'll use the datasets at Cancer Imaging Archive and download a study. I get a folder with some DICOM .dcm files. Trying to open one shows that they are stored as binary format. The DICOM standard tells us how to decode this.
I open with an online DICOM viewer. There are 92 images in the study I downloaded, and a bunch of meta data. Finding out exactly how the images are stored in the .dcm file is a bit tricker. It seems that .dcm files can include things like JPEG, TIFF, and raw pixel data, and you can use the metadata to decode each image. They can also contain raw data, or acquisition vales, that need to be reconstructed into the image.
You'll probably need a library like pydicom to be able to process the data
What other common file types exist?
There is another huge repository of medical imaging data called OpenNEURO that converts the .dcm files into their own Brain Imaging Data Structure (BIDS). BIDS is a standardized directory structure that stores the MRI image files as .nii files (Neuroimaging Informatics Technology Initiative). These files store the intensity of the MR signal at each of voxel in the image.
What about ultrasound data?
The raw data of ultrasound as measured by the device, is a time series of electrical signals caused by bouncing ultrasonic acoustic waves off of the tissues deep in the body. After image reconstruction, the resulting image shows the acoustic impedance at each cell or voxel. With a bit of post-processing, this data can be converted into wave speed at each point in the image.
Most of the time you'll find ultrasound data stored in .dcm files. Inside the .dcm file you might find JPEG or TIFF images. You might also find raw data in the form of time signals.
Is there ultrasound data of the brain?
A little bit.
Ultrasound imaging of the brain is only just recently becoming feasible with something called the Full Waveform Inversion (FWI) algorithm. So far, nobody has been able to create a clinically viable form of this technology, so we don't have that data.
However, neonatal ultrasound scans exist that image a fetuses brain in the womb, before the skull is fully developed. Also, cranial ultrasound for adults is possible, but only after the skull has been opened during brain surgery.
That data comes in 2D images (often .png files) and is often pretty blurry (see fig. 2)
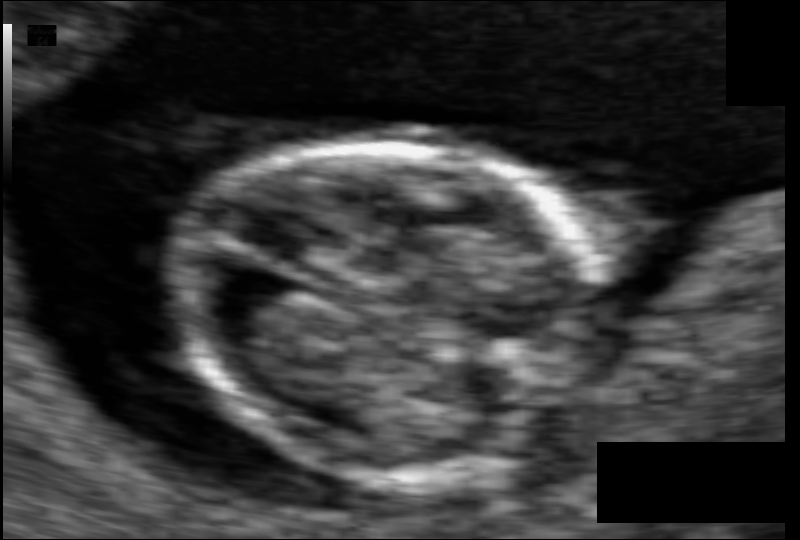
Figure 2: Neonatal ultrasound data from Alzubaidi et al [1]
How about 3D Models of the brain?
These exist, and they generally come from data from OpenNEURO. The .nii files are combined to make a 3D model.